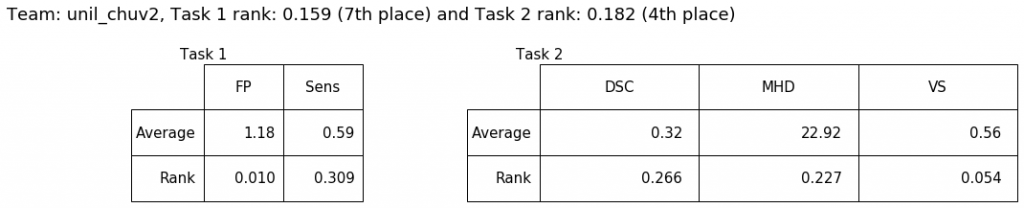
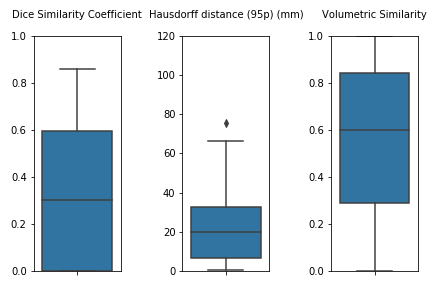
This is a further version of the original unil_chuv method submitted for task 1 at MICCAI 2020.
Changes relative to first submission:
- increased overlapping in the sliding window approach (before it was 0, now it’s 25%). This was probably the thing which had the biggest impact on sensitivity
- resampled all volumes to median voxel spacing
- removed FP smaller (in mm^3) than the smallest of all ADAM aneurysms
- increased the number of training 3D patches extracted for each subject, from 30 to 40
- Decreased the UNet threshold from 0.7 to 0.4
- used bias-field corrected volumes instead of original
- checked patients where registration was not correct: if it was wrong, I did not take the distances to the landmark points int account during the sliding window
